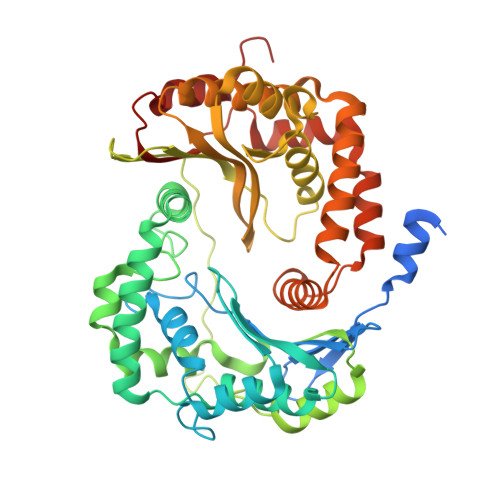
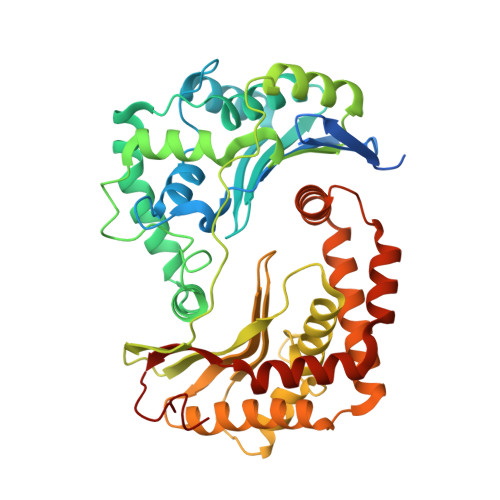
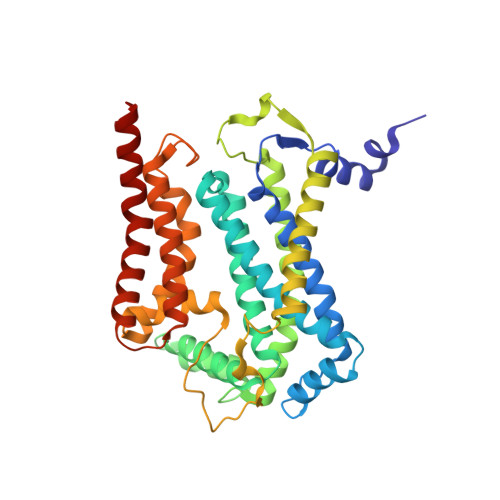
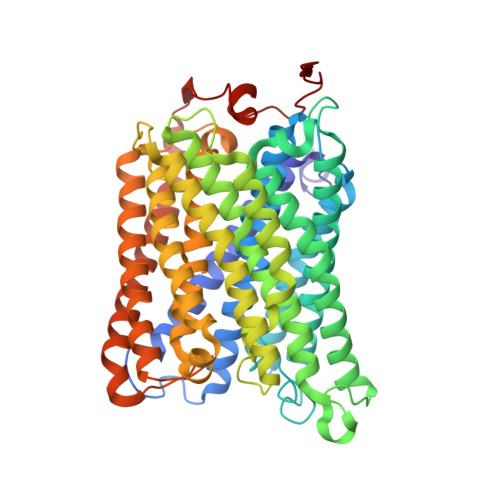
SCAF1 drives the compositional diversity of mammalian respirasomes.
Vercellino, I., Sazanov, L.A.(2024) Nat Struct Mol Biol
- PubMed: 38575788
- DOI: https://doi.org/10.1038/s41594-024-01255-0
- Primary Citation of Related Structures:
8PW5, 8PW6, 8PW7, 8RGP, 8RGQ, 8RGR, 8RGT - PubMed Abstract:
Supercomplexes of the respiratory chain are established constituents of the oxidative phosphorylation system, but their role in mammalian metabolism has been hotly debated. Although recent studies have shown that different tissues/organs are equipped with specific sets of supercomplexes, depending on their metabolic needs, the notion that supercomplexes have a role in the regulation of metabolism has been challenged. However, irrespective of the mechanistic conclusions, the composition of various high molecular weight supercomplexes remains uncertain. Here, using cryogenic electron microscopy, we demonstrate that mammalian (mouse) tissues contain three defined types of 'respirasome', supercomplexes made of CI, CIII 2 and CIV. The stoichiometry and position of CIV differs in the three respirasomes, of which only one contains the supercomplex-associated factor SCAF1, whose involvement in respirasome formation has long been contended. Our structures confirm that the 'canonical' respirasome (the C-respirasome, CICIII 2 CIV) does not contain SCAF1, which is instead associated to a different respirasome (the CS-respirasome), containing a second copy of CIV. We also identify an alternative respirasome (A-respirasome), with CIV bound to the 'back' of CI, instead of the 'toe'. This structural characterization of mouse mitochondrial supercomplexes allows us to hypothesize a mechanistic basis for their specific role in different metabolic conditions.
Organizational Affiliation:
Institute of Science and Technology Austria, Klosterneuburg, Austria.